Littman Lab


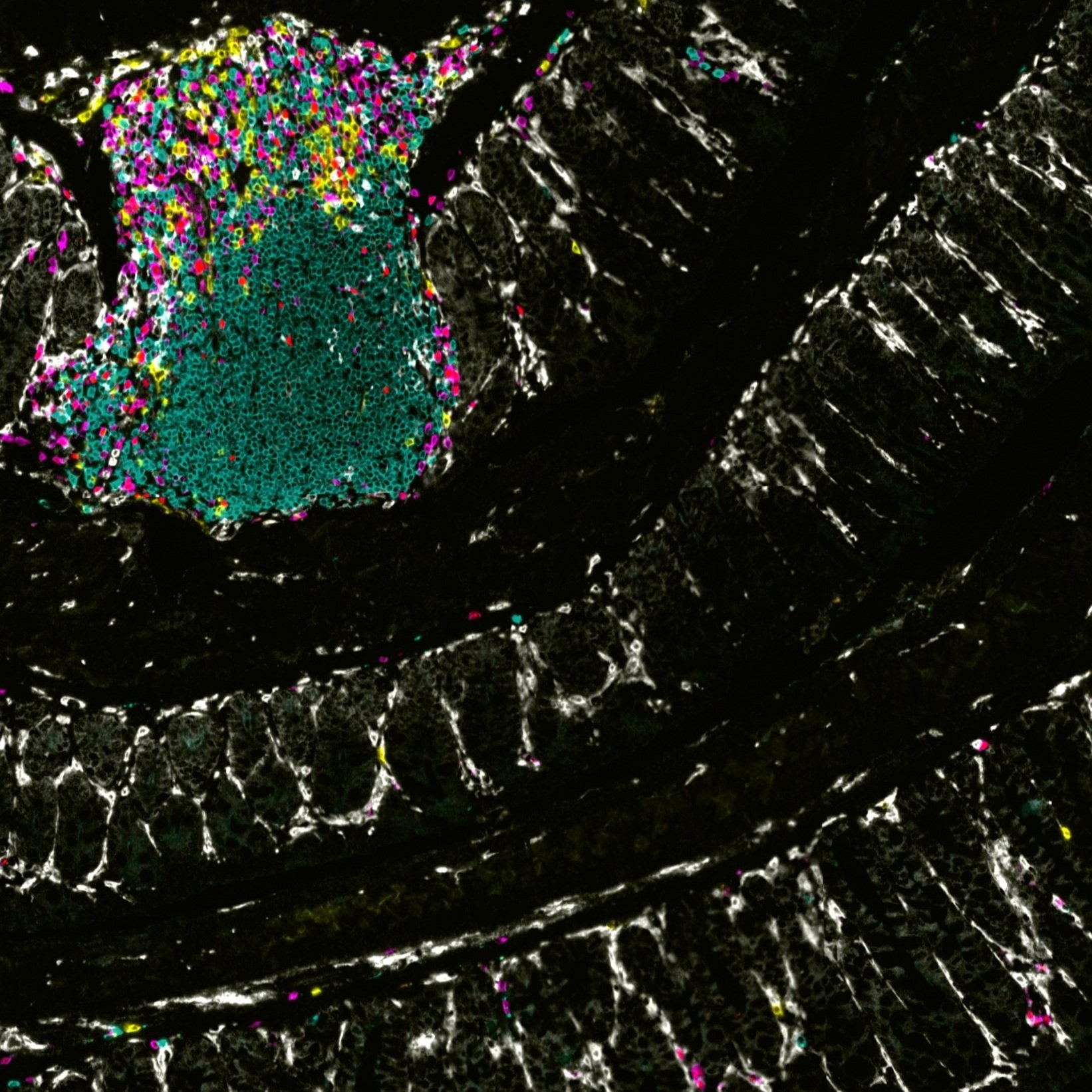

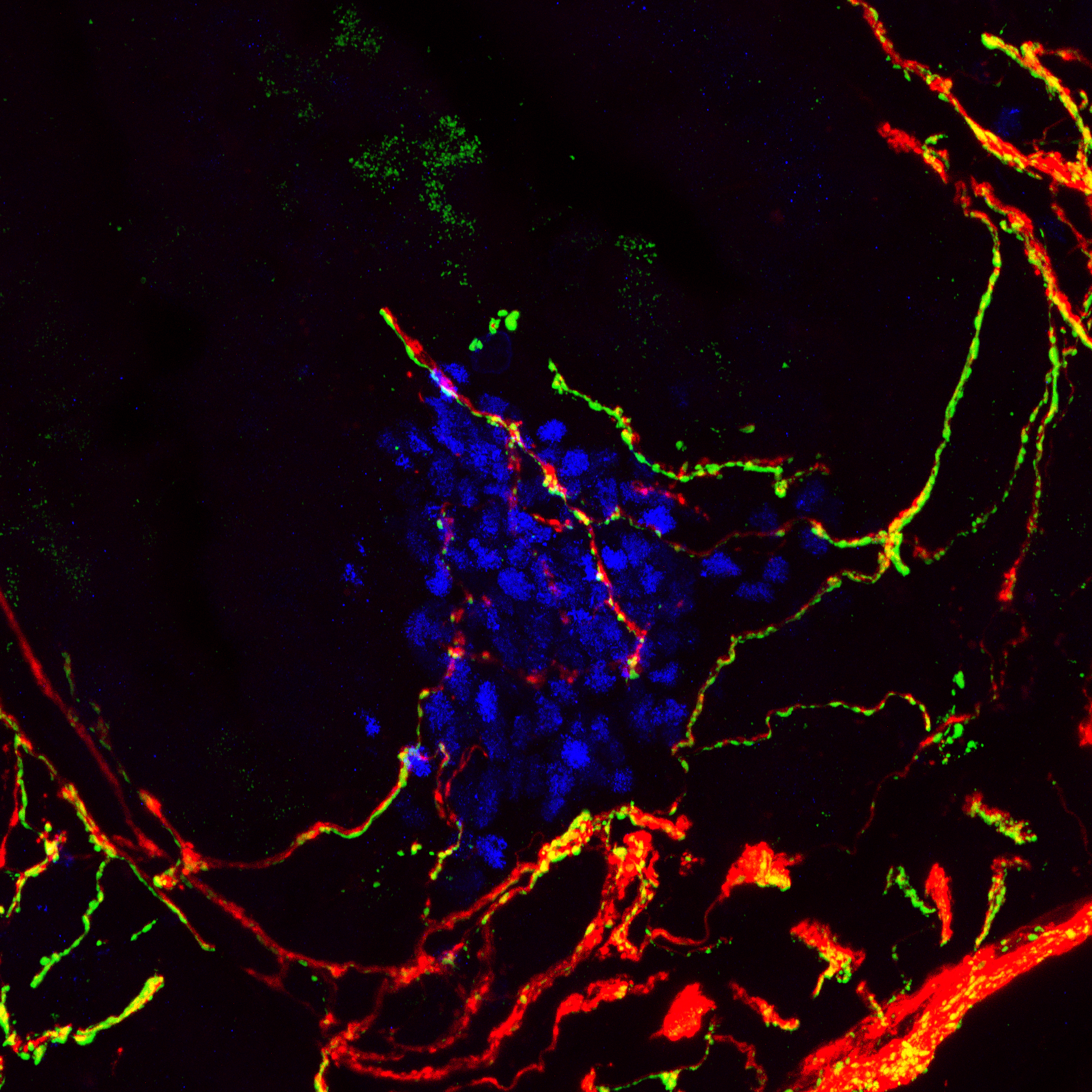





The vertebrate immune system protects the host from a multitude of potentially pathogenic microorganisms. Although this is commonly thought to be the principal role of the immune system, it is now recognized that its diverse components are deployed in numerous physiological processes, ranging from tissue repair and regeneration to pruning of neurological synapses to conveying information on nutrient availability and metabolic requirements. Disturbances in immune system functions can hence result in a wide variety of diseases, including not only those of the autoimmune and inflammatory variety, but also cardiovascular, metabolic, and psychiatric/neurodegenerative diseases. An additional layer of complexity is provided by the commensal microbiota, whose composition is linked to a wide variety of physiological functions, many of which are conveyed through its effects on the immune system. Our laboratory studies how information from the environment, including microbiota and metabolites, is relayed to cells of the immune system and how this is manifested in homeostatic processes as well as in pathological conditions.
